Cot curve is a technique used to measure the complexity of DNA or a genome. The DNA renaturation is analysed by this method.
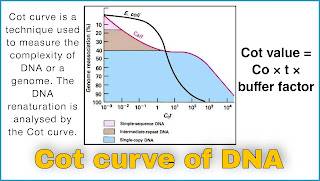
Cot curve of DNA
Genome complexity is a total length of different sequences of DNA. It can be measured by the renaturation kinetics of denatured DNA. Renaturation of DNA occurs because of complementary base pairing of DNA strands. Renaturation depends upon random collision of the complementary DNA strands.
This percentage of DNA is a ratio of Concentration (Co) of single stranded DNA (ssDNA) and the total concentration of DNA .
Along the X axis the product of initial concentration of DNA and the time (in second) taken for the reaction is taken. After plotting the X and Y axis the value comes out as Cot. So the curve is also called the Cot curve of DNA. At the time of calculating the Cot value, a buffer factor that accounts for the effect of cation is also taken into consideration.
Cot curve principle
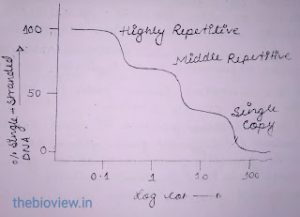
- The rate of renaturation of DNA is directly proportional to the number of times the sequences are present in the genome.
- Enough time is given so that all DNA that is denatured will reassociate or reanneal in a given DNA sample.
- The more the repetitive sequence the less will the time take for DNA renaturation.
- DNA concentration
- Renaturation time
- Temperature
- Buffer factor
- Viscosity
Read more from us in the topic Polymerase Chain Reaction
Cot value / Cot Equation
Degree of Renaturation after given time depends on Co and t.
Cot value = Co × t × buffer factor

Meaning of Co, t and buffer factor in cot equation
Co stands for nucleotide concentration in moles per liter prior to denaturation.
Read More about difference between chloroplast DNA and Mitochondrial DNA
COT Analysis
- The genomic DNA is first cut randomly into fragments about 1kb in length.
- Fragments are completely denatured by heating above the melting temperature. (Tm =temperature at which 50% of the DNA is melted)
- Fragments are allowed to renature(renaturation at a temperature about 10°C below the Tm).
Renaturation at different time intervals can be monitored by two ways:
- using Spectrophotometer(by taking absorbance at 260nm- as ssDNA absorbs light more efficiently than ds DNA (due to base stacking it has more constrained bases).
- using Hydroxylapitite chromatography(HC)- an adsorption chromatography, Hydroxylapitite (modified form of crystalline calcium phosphate) adsorb double stranded DNA as the positively charged calcium in hydroxylapitite will have more affinity towards double stranded DNA as dsDNA consists of more negatively charged phosphate groups than ssDNA.
FAQ related to Cot curve
What is the use of Cot curve analysis?
Analysis of a Cot curve gives data about genome size, the proportion of the genome contained in the single-copy and repetitive DNA components, and the kinetic complexity of DNA.
What is cot value and what does it measure?
Degree of Renaturation after given time depends on Co and t. Cot value = Co × t × buffer factor. Co = nucleotide concentration in moles per liter prior to denaturation. t = reassociation time in seconds. buffer factor= a factor based upon the cation concentration of the buffer.
What is Cot curves of DNA?
When the extent of renaturation of DNA is plotted against log Cot , this is called Cot curve.
